The ExFAB CSU Masters Scholars program provides CSU Masters and senior-level CSU undergraduate students the opportunity to participate in a 10-week paid summer internship in an ExFAB research laboratory located at either UC Santa Barbara or UC Riverside.
Applications are now open!
Program Information
Program Dates: June 8, 2026 – August 14, 2026
Application opens: January 9th, 2026
Deadline to Apply: March 2, 2026 @11:59PM
Estimated Date of Notification: Mid April 2026
Eligibility: Full-time CSU Masters or senior undergraduate students in a STEM major or area of study who have been accepted into a STEM-related CSU Master’s program for Fall 2026.
Eligible students must submit a completed application form before the application deadline.
CSU Master Scholar Host Labs
All applicants will have the opportunity to rank preferences on which lab they would like to visit, based on their interests and research.
UC Santa Barbara Labs
UC Riverside Labs

Ammonia-oxidizing archaea (AOA) are one of the most abundant groups of microbes on the planet, and can be found from the surface ocean to the hadal depths. Our lab is studying the adaptations AOA have for dealing with high hydrostatic pressure in the deep ocean, and how pressure alters their cellular chemistry and gene expression. Students will gain exposure to working with fastidious microbes, operating pressure chambers, microscopy and potentially transcriptomics and/or lipid analysis if time allows.
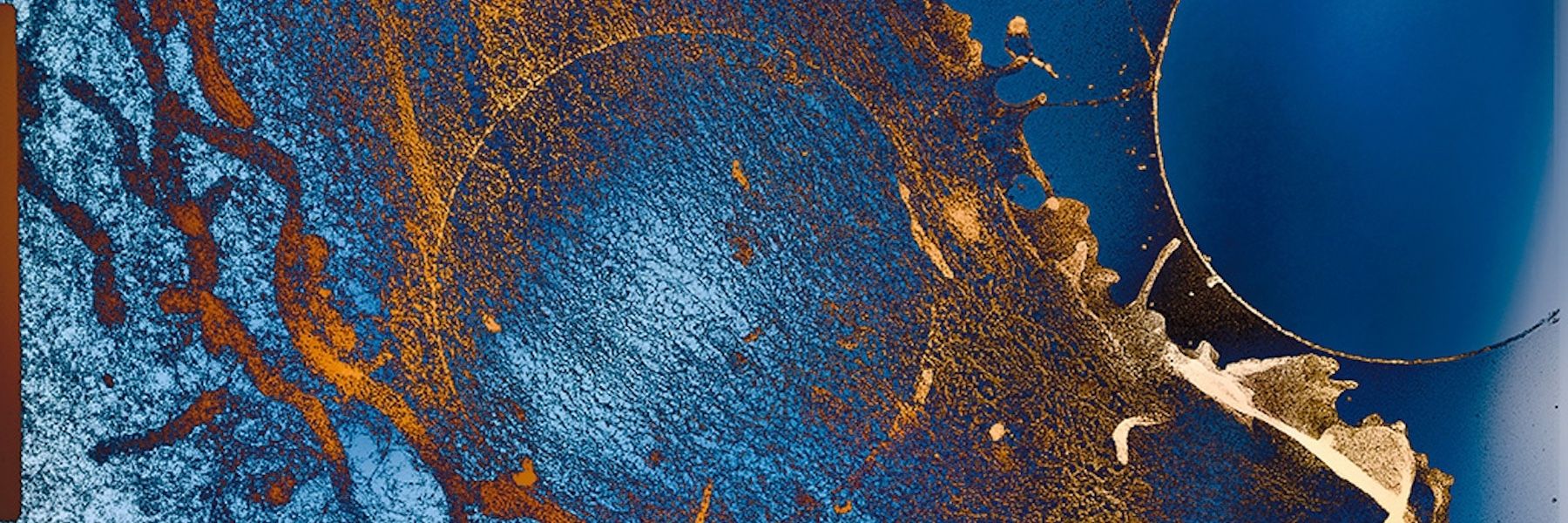
For a prospective CSU Master Scholars student, we would provide undergraduate-level foundational training in cryogenic Transmission Electron Microscopy (cryo-TEM), balancing hands-on and theoretical learning. The student would be introduced to grid handling, sample preparation, and negative-stain TEM. If microscope time can be supported, the student would be able to observe data collection and use the resulting dataset to learn the fundamental steps of image analysis.

This project will involve high throughput cultivation and screening of novel species of multicellular, biofilm forming phototrophic sulfur bacteria (purple sulfur bacteria). The student working on this project will have the opportunity to learn methods for anaerobic cultivation of diverse bacteria and preliminary genome analysis; and will work alongside ExFab scientists to design and implement a high throughput, AI-assisted screening and growth assay using ExFab's Cytation C10 automated confocal imaging system. This project is a part of our group's long term studies of a novel, natural symbioses between bacteria and informs questions regarding the "rules" of microbiome community assembly and the evolution of multicellularity.
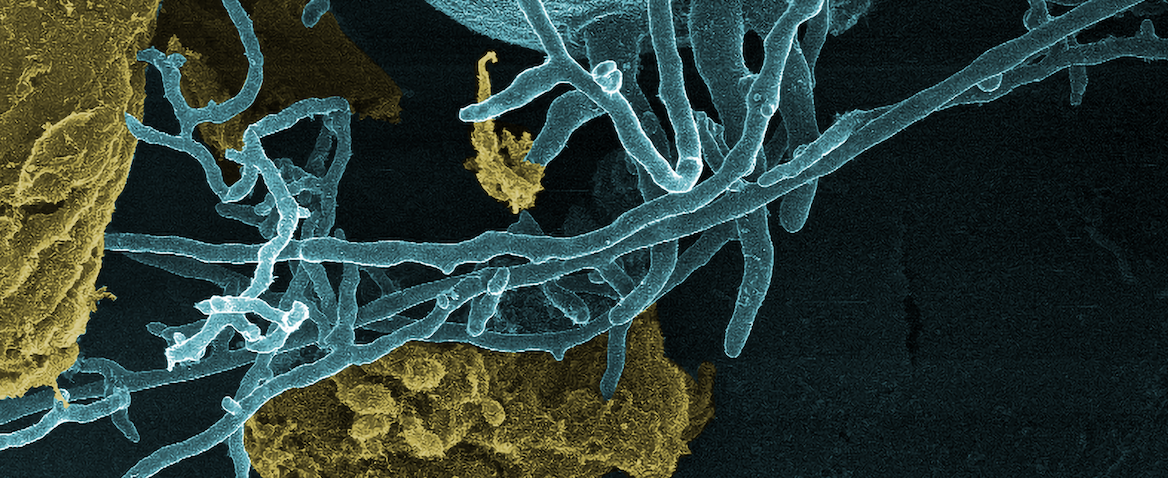
Our group develops tools to explore and engineer non-model microbes and microbial communities – especially for “anaerobic” microbes that live without oxygen. We combine classical microbiology with state-of-the-art approaches in -omics, genetic engineering, and high-throughput lab automation to investigate and reprogram microbial processes. As a Master Scholar participant, the student would be working with a mentor whose work focuses on growing anaerobic fungi and optimizing genetic engineering techniques to test protein function.
The Greene Lab is a collective of scientists interested in how biology performs reduction/oxidation (redox) reactions far from equilibrium in water, the universal solvent of life. The student would work on protein engineering of a redox-active flavoprotein for fundamental structure-function studies and applications in artificial electron bifurcation.
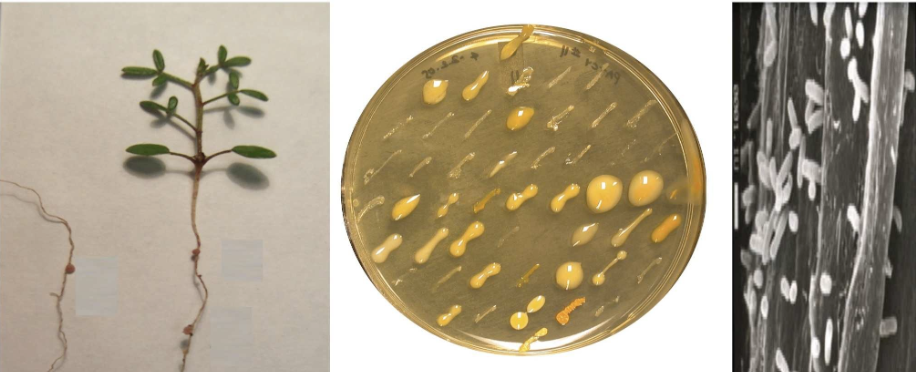
Symbiotic bacteria associate with plants and animals and provide critical services to these hosts. Despite the apparent benefits of these associations for the bacteria, these taxa often experience frequent evolutionary transitions between symbiotic and nonsymbiotic states. Using a combination of genomics, experimental evolution, and in vitro experiments, we study the evolutionary origins and stability of symbiosis between nitrogen-fixing bacteria and their plant hosts.
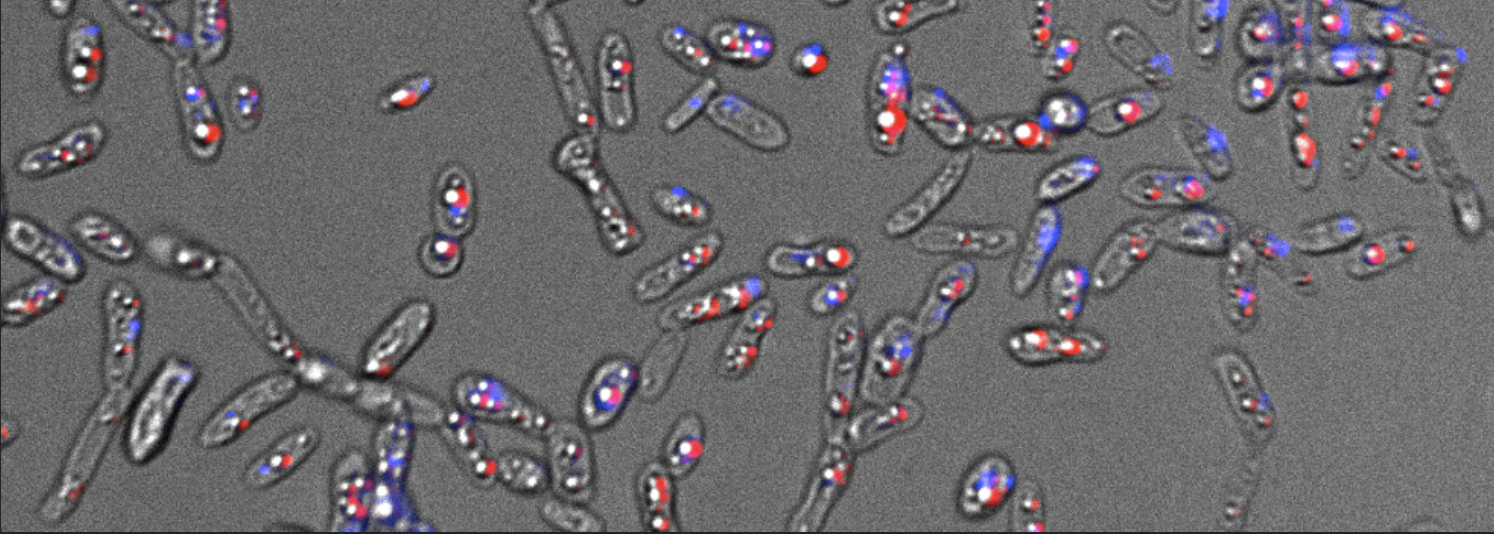
ExFAB seeks to develop automated workflows to enable the discovery, phenotyping, and genotyping of microbial isolates. This project seeks to establish a robust, high throughput workflow that links ExFAB’s phenotyping capabilities with genotyping via genome sequencing. A critical step in this pipeline is DNA extraction, which is traditionally accomplished in labor intensive bench top work. To automate DNA extraction, we will use ExFAB’s newly acquired QiaCube HT, a robotic liquid handler with associated thermocycler and vacuum manifold. Using a strain library of the red yeast Rhodotorula, students assigned to this project will develop and validate DNA extractions.
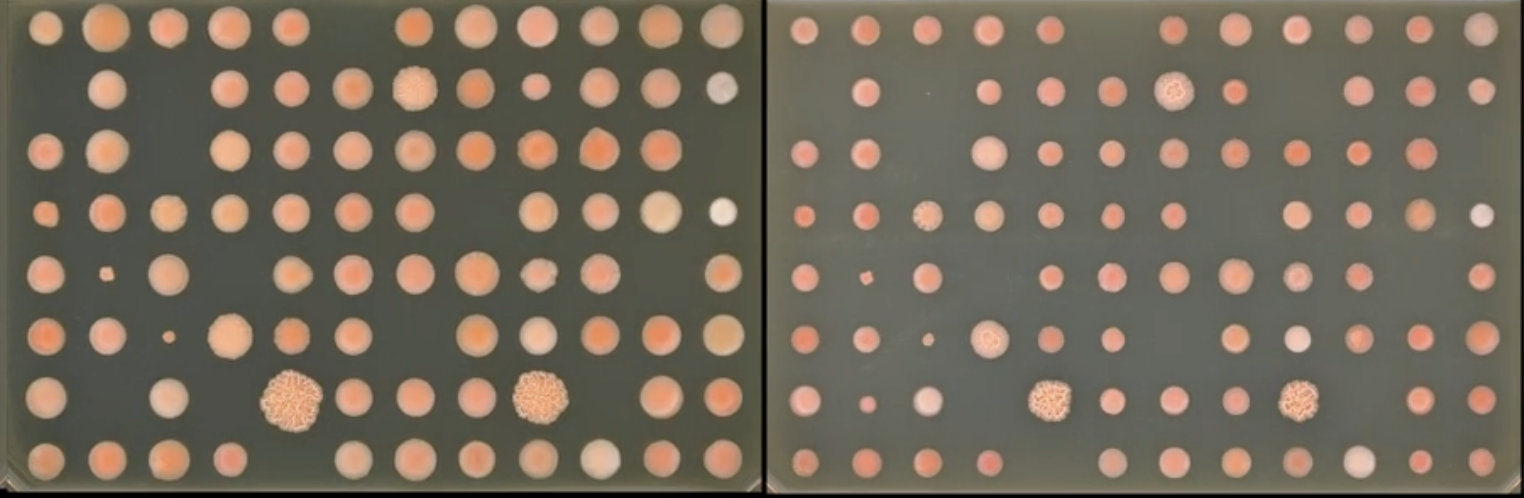
Rhodotorula genus of red basidiomycete yeasts that can be found growing in diverse environments from shower curtains to glaciers to cheese to heavy metal and radioactive contaminated environments. Its resilience suggests it has potential for bioremediation applications. Our goal is to conduct high-throughput phenotyping of a global collection of annotated and sequenced strains to identify genes that confer tolerance to these contaminated and stressful environments. Using ExFAB’s S&P pinner and imager platform to collect growth rates on solid media and apply the software Phenotypic, developed at ExFAB, to calculate growth rate information for use in genome-wide association studies.
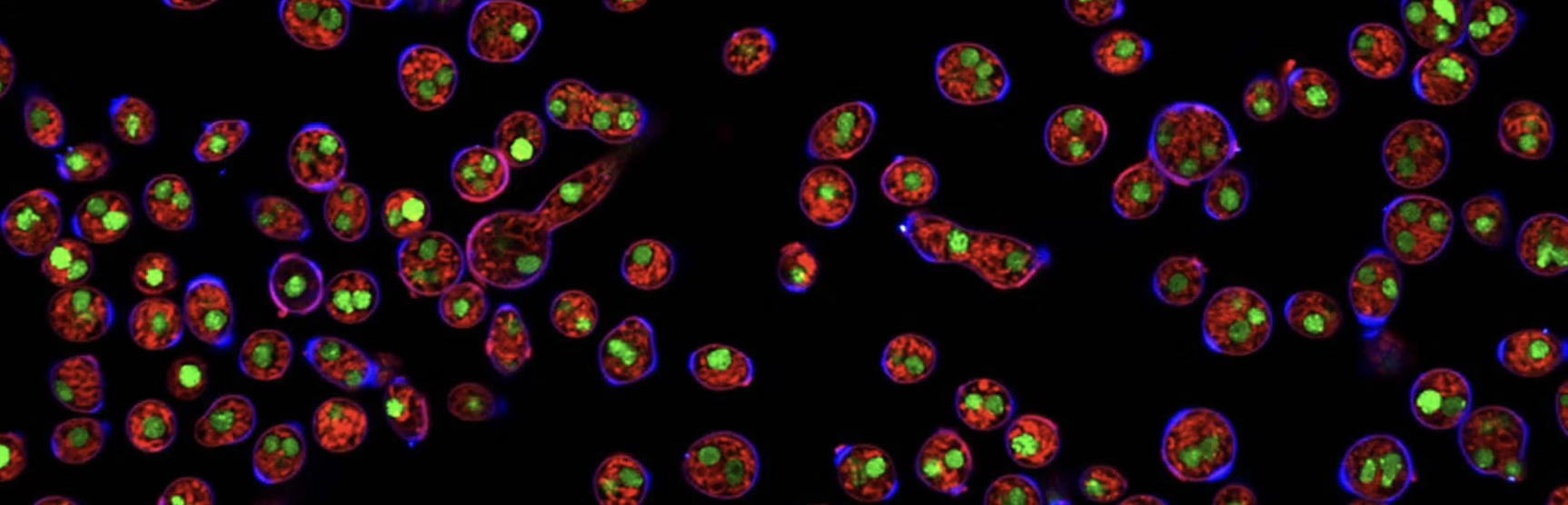
Graduate student Monique Quinn has been analyzing growth rate phenotypes for a large group of gene knockout mutants in the filamentous fungus Neurospora crassa using the imaging equipment and robotics in the UCR ExFAB facility. The mutants include all available strains lacking G protein coupled receptor (GPCR) genes. GPCRs are important for environmental sensing and signal transduction in all eukaryotic organisms. The MS student will analyze ~200 knockout mutants lacking genes encoding small secreted proteins (SSPs; 100 amino acids or less), as these are candidate ligands for GPCRs. The phenotypes of the GPCR and SSP mutants will be compared and putative receptor-ligand partners identified for further study.
Please note that all applications must have the following documents attached:
1. Personal Statement
- Describe your long-term career goals.
- Describe the experiences that shaped your interest in STEM.
- Discuss any other relevant information that you feel the selection committee should take into consideration when assessing your application.
- Describe your interest in the ExFAB CSU Masters Scholars program, including what you would like to gain from the experience.
2. Research Statement
- Please provide a brief description of any research experience you have had in the past and your role in the research project.
- Discuss any other relevant information that you feel the selection committee should take into consideration when assessing your experience in research.
3. Your C.V. (curriculum vitae)
4. Your Unofficial CSU Transcript
5. Three (3) Letters of Recommendation.
Letters of Recommendation should be on a letterhead. They should be from a science faculty member, investigator or employer who is qualified to comment on your attributes and can comment on your potential for success as a researcher. Please have the letters of recommendation submitted by the recommender through the “Submit a Letter of Recommendation.”
Current Scholars



